Amazon Eks Based Genomics Data Analysis Platform With Aws Primary Storage Solutions
Name and Sector of Client:
The healthcare company is a giant in Genomics research Field and Healthcare sector spanning services pan-India. Their expertise includes:
- Advanced genetic data analysis capabilities to urgent health challenges, developed in response to diseases.
- Partnerships with local hospitals across Hyderabad, enabling wide-reaching impact in healthcare research.
- Large-scale genetic information processing using a sophisticated data analysis platform built on Amazon EKS.
- Handling of complex genomic workflows, from raw sequencing data to final analysis outputs.
- Multi-institutional collaboration, analyzing genetic data from multiple healthcare institutions across India.
Data Sources of Client:
Due to this broad aspect of their research model, they gather huge volume of data spanning these listed sources & beyond.
The org pulls genomics data from External Research Databases:
- Public genomic databases (e.g., NCBI, EBI)
- SRA (Sequence Read Archive) for raw sequencing data
- Human Genome Diversity Project (HGDP) for population genetics studies.
Partnered Hospitals/Healthcare Institutions:
- Electronic health records (EHRs)
- Hospital information systems
- Laboratory information management systems (LIMS)
Data type and Volume:
Raw Sequencing Data (FASTQ files):
- Category: Unprocessed genomic data
- Amount: Can vary widely, but typically 10-30(approx) GB per human genome
- Daily ingestion: Depends on sequencing capacity, could be 1-10 full genomes per day
Aligned Sequence Data (BAM files):
- Category: Processed genomic data
- Amount: Likely or slightly larger than raw data, 10-50 GB (approx)per genome
- Daily ingestion: Corresponds to raw data processing, 1-10 per day
Clinical Data:
- Category: Patient information, medical history, test results
- Amount: Much smaller, typically <5 MB per patient
- Daily ingestion: Could be hundreds of records, depending on hospital partnerships
These file types often represent different stages in genomic analysis procedure. The process starts with FASTQ files and by aligning them to a reference genome to produce BAM files, and then perform various analyses on the aligned data.
Current Total Monthly Data Ingestion: Approximately 15-20 TB per month
All these data from various sources are ingested in Amazon EKS by using APIs provided by the 3 rd parties.
Applications running on Amazon EKS:
- FastQC a quality control tool for checking the quality of raw sequencing data (FASTQ files)
- Trimmomatic CLI tool for trimming and filtering low-quality reads or text formats
- BWA (Burrows-Wheeler Aligner) for aligning DNA sequences to a reference genome
- Bowtie2 tool used for aligning for DNA and RNA sequences.
- Picard for manipulating and analyzing SAM/BAM files.
Analysis process:
- Raw data (FASTQ files) would be read from DB and processing via the tools starts.
- Intermediate results (e.g., aligned BAM files, variant calling formats) would be stored in Amazon EFS for quick access by other pods which would be used by subsequent steps in the pipeline.
- Final processed data (json, yaml, csv, HTML, BAM) would be generated and potentially stored back in long-term storage.
Challenges faced by the client:
- Critical requirement forICMR-compliant and Indian regulations for data accessibility across multiple Availability Zones (AZs), ensuring continuous availability and enabling disaster recovery capabilities for sensitive genomic information in accordance with healthcare regulations.
- Necessity to streamline storage provisioning and management, significantly reducing administrative overhead and allowing researchers to focus exclusively on genomics work.
- Requires a cost-optimizationof the environment that aligns storage and compute resource usage with actual needs, eliminating unnecessary expenses.
- Single-region storage violates industry standards for data redundancy and disaster recovery, risking critical data loss and regulatory non-compliance.
- Inconsistent Backup and retention practices implemented by the organization possess to meet regulatory requirements for genomic research data preservation, jeopardizing research integrity and legal compliance.
- The researchers/org users were given certain bucket level permissions which leads to the users access to all the data present in the bucket.
- No advance level permissions with granularity were present in the environments for the users to access the data from Amazon S3which raised the risk for the users to access certain data that are not to be accessible by them.
- The environment suffered from significant over-provisioning and underutilizationof resources. This resulted in unnecessary costs and inefficient resource allocation and calls for optimization requirement procedure.
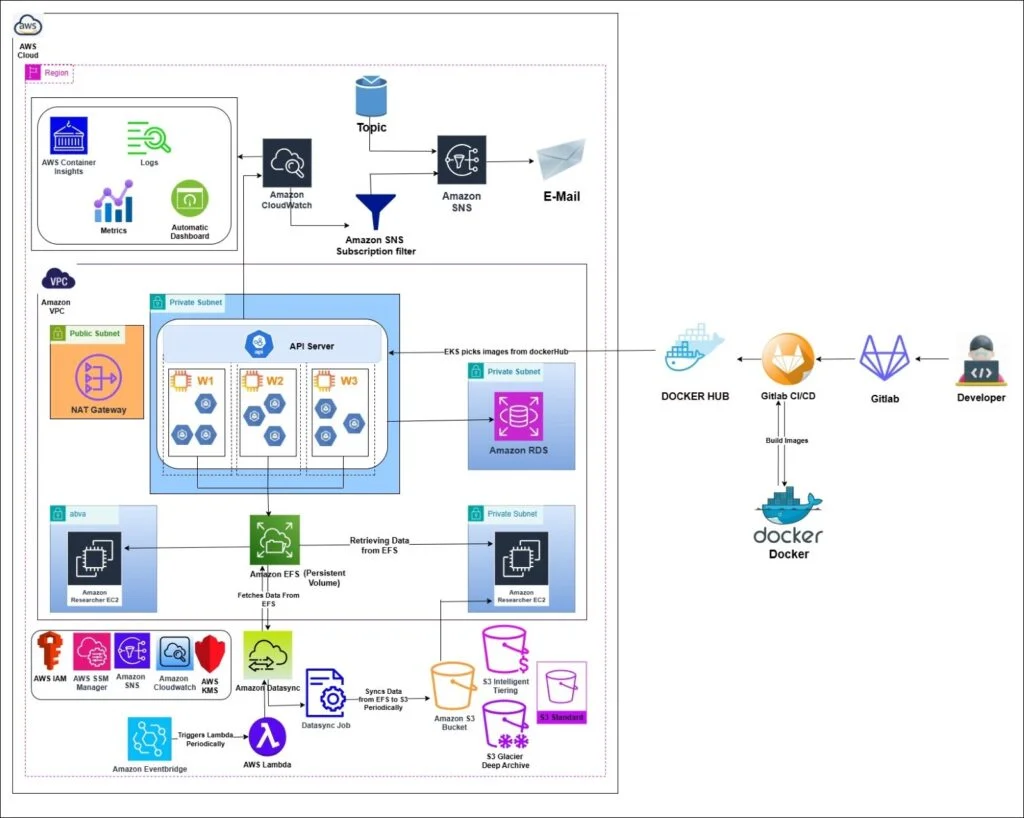
Proposed Solution and Architecture:
Amazon EFSis utilized to store various types of genomic data crucial for research and analysis. This includes large volumes of raw sequencing data in FASTQ files generated by sequencing machines, which need to be accessed quickly and concurrently by multiple researchers and pods for quality control and preprocessing.
During sequence alignment, worker nodes and pods in the Amazon EKS cluster process these raw data files using tools like BWA and Bowtie, producing aligned BAM files.
These BAM files, along with intermediate results and reference genomes, must be immediately available to other nodes and pods for further processing and analysis, ensuring data consistency and eliminating the need for data duplication or transfer.
Type of Data stored in Amazon EFS and Amazon S3:
Data kept in Amazon EFS:
- Active research data: Currently processed genomic sequences and ongoing analyses.
- Intermediate results: Temporary files generated during analysis pipelines.
- Frequently accessed reference data: Common reference genomes or annotation files which are frequently accessed by the Researchers.
- Shared scripts and tools: Custom analysis scripts and frequently used bioinformatics tools.
Data transferred to Amazon S3 periodically via Amazon DataSync:
- Completed analysis results: Finalized variant calls, gene expression profiles, or assembled genomes.
- Raw sequencing data: Original FASTQ files after initial quality control and preprocessing.
- Large, processed datasets: Aligned BAM files or variant call format (VCF) files from completed analyses.
- Periodic snapshots: Hourly backups of critical research data for disaster recovery.
These kinds of data cannot be stored in Amazon EFS for an extended period due to the high cost and potential performancedegradationover time, especially as the volume of data grows. Therefore, they are moved to Amazon S3 via Amazon DataSync job for more cost-effective, scalable, and durable long-term storage.
Dynamic Provisioning Implementation for Streamlining storage provisioning
Why choose Dynamic Provisioning over Static Provisioning?
Dynamic provisioning allocates storage resources precisely when they are needed, based on the application's requirements. This avoids the inefficiencies of pre-allocating fixed storage sizes with static provisioning. Dynamic provisioning allows for seamless adjustment of storage allocation without downtime or manual resizing optimizing the whole volume provisioning.
In this scenario the uncertainty of the amount of volume that is required to be provisioned lead to the perfect use of Dynamic Provisioning. DP uses Kubernetes StorageClassesto automatically create Persistent Volumes (PVs) when Persistent Volume Claims (PVCs) are made.
For example, if an application needs a 100GB volume, a PVC can be created specifying this size, and Kubernetes will automatically provision a new PV of 100GB using the defined StorageClass. This eliminates the need for manual creation and management of PVs, reducing administrative overhead and ensuring that storage is always available when needed without manual intervention.
Ancrew Installed the Amazon EFS CSI Driver in theAmazon EKS cluster, which automated the installation of necessary utilities and manages Amazon EFS volumes through Amazon EFS CSI Driver Add-on and by utilizing the add-on the Dynamic provisioning solution was implemented.
Why introduce Amazon EFS?
Amazon S3: Amazon S3 is an object storage service that does not support POSIX compliance file system semantics. It is designed for storing and retrieving large objects but does not support the fine-grained control and real-time file access needed for computational tasks that require standard file operations.
Amazon EBS: Amazon EBS is POSIX compliant but cannot be used in this case as we cannot mount it on multiple Amazon EC2s (worker nodes). Also, Amazon EBS are AZ specific, and it cannot share data out of AZ.
Amazon EFS: Whereas AmazonEFS is POSIX compliant, allowing it to support the standard file system semantics required for bioinformatics tools.
Amazon EFS can be mounted on multiple Amazon EC2 instances across different AZs, providing shared, scalable, and distributed file storage that ensures data redundancy, high availability, and seamless access to shared data across your Amazon EKS cluster.
This makes it ideal for the high-performance, real-time data processing needs of your genomics analysis platform.
AmazonS3 enhancements suggested by Ancrew:
Data coming into Amazon S3 is categorized and then stored in the buckets with certain tagging.
The users were given certain permissions based on their requirements but only on bucket level but not on object level.
This increases the risk of the users to see all the different kinds of data present in the folder.
Ancrew suggested to implementtag-based objects and this lead to the users of accessing the required bucket, required folder and along with that the required objects only and thus implementing the advance level accessing control over the users, way deep till the objects.
Ancrew team wrote AWS Lambda function to tag the new uploads happening to Amazon S3 buckets through Amazon Datasync.
This AWS Lambda function runs after Amazon Datasync job is successful and is triggered by Amazon Eventbridge.
This AWS Lambda function with permissions like PutObjectTagging, ListBuckets,etc,extracts bucket name and the folder prefix in which data is stored in Amazon S3.
The naming convention of the objects were unique like for processed_genome (pg), raw_sequence (rs), etc.
Using the unique parameters such as naming convention and prefix of the destination folder of the object's tagging logic has been built and the objects were tagged and uploaded to Amazon S3 bucket.
How Ancrew ensured the tagging of the already existing data?
This is done by using AWS Lambda function which checks if the tagging on the objects is present or not in the certain bucket for which it is written for and if not then it tags the objects based on the parameters like project_name, naming convention, etc.
How Ancrew monitored the implemented tagging mechanism?
Ancrew has written a AWS Lambda function which runs in daily to check if the tagging is done on the buckets or not.
If any untagged objects were found, an Amazon SNS alert with the list of untagged objects is sent to the team and immediate action is taken on it.
Transfer Data from AMAZON EFS to AMAZON S3 Using Datasync
- Amazon DataSync efficiently transfers data from Amazon EFS to Amazon S3 at regular intervals, ensuring cost-effective long-term storage of processed genomic data.
- AWS Lambda function automates the Amazon DataSync job initiation, minimizing operational overhead and reducing the need for manual intervention.
- Amazon EventBridge triggers the AWS Lambda function every ½ hour, maintaining a consistent data transfer cadence that aligns with research workflows and data generation patterns.
- By moving data from Amazon EFS to Amazon S3, the solution optimizes storage costs while maintaining data accessibility for future reference or analysis.
- This setup seamlessly handles increasing data volumes, supporting the organization's growth and evolving research needs without requiring significant infrastructure changes.
- Regular data transfers facilitate data lifecycle management, aiding in meeting retention policies and regulatory requirements specific to genomic research.
Script:
Architecture:
Security:
- Encryption of Amazon EBS volumes by using AWS KMS for implementing security on data stored on Amazon EBS.
- Ancrew suggested to used Multi-regional AWS KMS keys to reduce the overhead for managing multiple keys for single volumes.
- Data transfer between Amazon EFS to Amazon S3 via Amazon Datasync by using TLS/SSL encryption.
- Configured security groups to allow NFS traffic (port 2049) between worker nodes and Amazon EFS mount targets.
- Updated the Amazon EFS file system policy to allow both elasticfilesystem:ClientWrite, elasticfilesystem:ClientRead, and elasticfilesystem:ClientMount actions.
- Encrypted the data at rest when present in Amazon EFS.
- Encrypted data in-transit by using Amazon EFS policies.
"Enforce in-transit encryption for all clients"
Monitoring:
Implemented container insights to get performance insights through metrics for the Amazon EKS pods.
Various important metrics like pod restarts, CPU/RAM utilizations of the nodes were monitored through Amazon CloudWatch automatic dashboards created while deploying Container Insights on Amazon EKS cluster.
Utilized Amazon CloudWatch metrics for monitoring of Amazon EFS such as DataReadIOBytes, PercentIOLimit, PermittedThroughput, etc to gauge the performance of Amazon EFS.
Similarly, Ancrew used cloudwatch metrics for monitoring Amazon S3 such as Request metrics, Replication metrics, etc.
Some important metrics utilized for Datasync were BytesTransferred, BytesPreparedSource, BytesWritten, etc.
Logging:
Suggested Enabling API server logging for Amazon EKS.
Implemented container insights where all the pod logs were transferred to Amazon CloudWatch logs.
Helped in central logging of all the pods, nodes, Amazon EKS components present in the environment.
Alerting:
Ancrew suggested using Amazon SNS and utilizing topics to send customized alerts based on subscription filters setupd on Amazon EKS to reduce response time of an error interruption events.
Alerts for whenever the Amazon Datasync job is triggered by AWS Lambda and this alert is sent to the concerned personnels.
Once the Amazon Datasync job is done a Amazon SNS notification is again sent to the team.
Implementation of Alerting when the AWS Lambda Cleaning Job start or is done.
Backup and Disaster Ready mechanism:
Cross-region replication for taking backupsof Amazon S3 using Amazon S3 cross-region replication.
This Amazon S3 replication setting once setupd, worked 24/7 continuously, to automatically replicate new objects added to the source bucket to the destination bucket in the secondary region.
For Amazon EBS volumes AWS backup was used to send the snapshot from ap-south-1(Mumbai) to ap-south-2(Hyderabad).
The backups were set to be taken dailyafter Business hours.
AWS Backupwas configured to automatically delete the snapshots after 1 weekthus managing the lifecycle of your backups without need for additional scripts or manual intervention.
Cost Optimization Implementations and Automations:
Ancrew provided AWS Lambda scripts for cost-optimization in Amazon EBS which does the work of deleting idle unattachedAmazon EBS volume resources.
These scripts were utilized monthly which provided the actual unattached volumes a window period of 1 month, if any necessary data is present.
AWS Lambda scripts were written in python for Amazon EC2s after Testing hours.
The data transfer job fromAmazon EFS to Amazon S3 is done throughout the day and during non-Business hours the cleaning of the temporary data becomes a necessity to maintain the performance of the Amazon EFS.
For this a data cleaning AWS Lambdafunction is triggered weeklyat specific time by setting a cron job under Amazon Eventbridgeto clean the data from Amazon EFS.
The notificationof this is sent out to the concerned personnels and the AWS Lambda does its work of cleaning.
Once the cleaning is done another Amazon SNS notificationis sent out to let the concerned team know about the completion of the event.
This leads to an allowance of a weekly retention period for the temporary data and cleans the data to free Amazon EFS of unnecessary temp or intermediate results data.
Factors and Outcomes:
- By implementing Amazon EFSwithAmazon EKS worker nodes in across multiple Availability Zones (AZs), the platform ensured continuous data access and enhanced fault tolerance. This setup provided resilience against potential outages and maintained high availability of data.
- Dynamic provisioningplayed a crucial role in optimizing costs by ensuring that the organization only paid for the Amazon EFS resources it needed at any given time. This approach avoided the cost of over-provisioning while still maintaining the necessary performance levels for intensive computational tasks.
- The solution dynamically provisioned Amazon EFS storage as needed, thereby eliminating the need for manual storage management. This automation allowed researchers to focus entirely on genomics analysis without the overhead of managing storage resources.
- Security was kept in check with all data being encrypted both in transit and at rest. Secure access controls were meticulously managed through Amazon EFS policies and security groups, safeguarding sensitive genomic data against unauthorized access.
- The architecture was designed to be highly scalable, supporting the easy expansion of AWS resources in storage. This scalability is essential for accommodating the ever-growing datasets and increasingly complex analyses typical in genomics research.
- The ReadWriteMany access mode of Amazon EFS was particularly beneficial, enabling multiple pods across different nodes to simultaneously read and write data. This capability fostered collaboration among researchers, allowing them to work together seamlessly and efficiently.
- AWS Backup was used to ensure seamless and reliable data protection for the Bio company's critical analysis data stored on Amazon EBS. By scheduling daily backups, it provided assurance against data loss, maintaining integrity during intensive genomics analysis.
Conclusion:
Ancrew provided a robust, cost-effective, and secure infrastructure that facilitated advanced genomics research by automating storage provisioning, ensuring high availability and fault tolerance, optimizing costs, and enabling collaborative data access.
Ancrew aided in addressing the pain-points of the organization in storage areas while maintaining compliance requirements and security in check.